| Version 13 (modified by ajj, 8 years ago) (diff) |
|---|
Exercise 9 - SANS Data Analysis using SasView
Introduction
This exercise will introduce you to analysing SANS data using geometrical models in SasView. You will first look at how different shapes produce different scattering patterns, and how the model parameters affect the scattering pattern. You will then load some real SANS data and attempt to fit models to the data in SasView.
The exercise is divided into 3 sections:
Before beginning the exercise, you must first ensure that SasView is installed. If you have not done so already, follow these installation instructions.
Tasks you should perform are show as:
TASK: Do this thing
1. Familiarisation with SasView
TASK: Start SasView. The application should open and look something like the images below.
The SasView user interface contains 4 main areas:
- The Data Explorer
- This is where data is loaded and can then be plotted or sent to the various types of analysis.
- Models not associated with data (called "Theories" in !Sasview) can be plotted and converted to datasets.
- The Analysis Panel (which defaults to showing Fitting)
- This is where you do the work of analysing data or generating theories
- SasView currently supports four analysis tools:
- Fitting - for theory generation or model fitting to 1D and 2D SANS, SAXS, or SESANS data
- P(r) Inversion - for converting I(Q) to P(r)
- Invariant - for calculating the scattering invariant from a 1D data set
- Correlation Function - for performing a correlation function analysis of a 1D data set
- The plot windows (which appear when something is plotted)
- The menus, toolbar, and status area.
The capabilities of SasView are described in more detail in the application documentation with links to the relevant parts of the documentation available as "Help" buttons in each part of the GUI.
TASK: Briefly familiarise yourself with SasView panels, menus and documentation. Try changing to different analysis tools.
2. Exploring geometrical models
3. Fitting SANS data
This part of the exercise will use real SANS data taken from a study of surfactant self assembly in deep eutectic solvents (DES).
TASK: Restart SasView
Before starting this part of the exercise, you should have a clean SasView instance. Quit SasView and restart it.
Background information
Deep eutectic solvents are a class of ionic liquids formed from a hydrogen bond donor and a halide salt. At a certain mixture ratio, the eutectic mixture, the melting point is significantly depressed to values below room temperature.
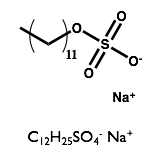
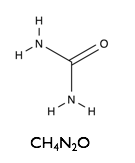
Here we will examine the self-assembly of a surfactant, sodium dodecyl sulfate (SDS) in the deep eutectic solvent formed from a 1:2 molar ratio mixture of choline chloride and urea.
| Sodium Dodecyl Sulfate | Choline Chloride | Urea |
 |  | 
|
TASK: Download the SANS data : TartuSasViewTutorialData.zip and unzip the file in a known location on your filesystem. Note where you have placed the data.
Loading and Plotting the data
You will now load the data into SasView and make a plot in order to visually inspect the scattering curves.
TASK: Click on the "Load Data" button in the Data Explorer
Locate the folder where you placed the data, select all the files in that folder and click "Open" in the dialog.
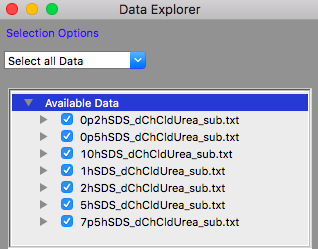
The Available Data section of the Data Explorer should look something like:
TASK: Plot the loaded data
Make sure that all the datasets have check marks next to them in the Available Data section of the Data Explorer, as shown above.
Click the "New Plot" button in the Data Explorer.
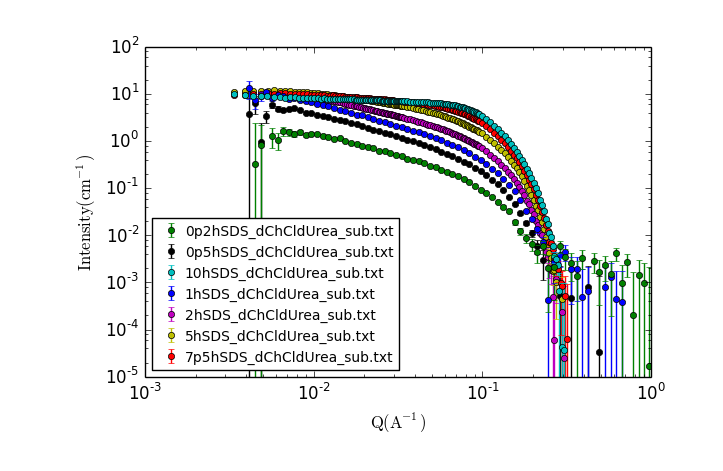
A new window should appear with a plot of the data that looks something like:
TASK: Examining the Data.
Visually inspect the data, zooming in and making additional plots as needed.
- What trends do you notice?
- What can you say about the possible solution structure from looking at the data?
Fitting the data
TASK: Fitting the lowest concentration data.
Select the lowest concentration data only in the data explorer by ensuring only 0p2hSDS_dChCldUrea_sub.txt has a check mark next to it and click “Send to" fitting.
- Select the model for the structure you predicted for this dataset.
- How does it compare to the data?
- Fill in parameters you know and adjust the others to see how close you get to the data.
- Select parameters to fit and run the fit by clicking "Fit" at the bottom of the fitting panel.
- Do you get a good fit?
- Are the parameters you get physically reasonable?
Attachments (18)
-
tartu_choline.png
(10.8 KB) -
added by ajj 8 years ago.
Structure of choline chloride
-
tartu_sds.png
(12.5 KB) -
added by ajj 8 years ago.
Structure of SDS
-
tartu_urea.png
(10.9 KB) -
added by ajj 8 years ago.
Structure of Urea
-
tartu_des.png
(16.7 KB) -
added by ajj 8 years ago.
A deep eutectic
-
TartuSasViewTutorialData.zip
(29.7 KB) -
added by ajj 8 years ago.
Tartu SasView? Tutorial Data
-
MicelleStructureinDeepEutecticSolvents.pdf
(3.5 MB) -
added by ajj 8 years ago.
PCCP Paper on Surfactants in DES
- tartu_loaddata.png (29.5 KB) - added by ajj 8 years ago.
- tartu_dataplot.png (69.7 KB) - added by ajj 8 years ago.
- tartu_sld.png (11.0 KB) - added by ajj 8 years ago.
- tartu_datafileslist.png (33.8 KB) - added by ajj 8 years ago.
- tartu_fitpage1_1.png (60.7 KB) - added by ajj 8 years ago.
- tartu_fitpage2_1.png (64.2 KB) - added by ajj 8 years ago.
- tartu_cylinder.png (23.1 KB) - added by ajj 8 years ago.
- tartu_sphere.png (24.2 KB) - added by ajj 8 years ago.
- tartu_sasviewmac.png (158.7 KB) - added by ajj 8 years ago.
- tartu_sasviewwin.png (55.3 KB) - added by ajj 8 years ago.
- MicelleStructureinDeepEutecticSolventsSupplementaryInformation.pdf (3.9 MB) - added by ajj 8 years ago.
- kemm37_volumefraction_sld.png (122.6 KB) - added by ajj 7 years ago.